Estimate uncertainty
This tutorial shows how to estimate uncertainty measures on the model predictions. The uncertainty measures implemented in
ivadomedare detailed in Technical features.An interactive Colab version of this tutorial is directly accessible here.
Download dataset
A dataset example is available for this tutorial. If not already done, download the dataset with the following line. For more details on this dataset see One-class segmentation with 2D U-Net.
# Download data ivadomed_download_data -d data_example_spinegeneric
Configuration file
In this tutorial we will use the configuration file:
ivadomed/config/config.json. First off, copy this configuration file in your local directory (to avoid modifying the source file):cp <PATH_TO_IVADOMED>/ivadomed/config/config.json .Please open it with a text editor. The configuration file will be modified to be the same as the one used for Technical features. As described in the tutorial One-class segmentation with 2D U-Net, make sure
path_datapoint to the location of the dataset. The parameters that are specific to this tutorial are:
path_output: Location of the directory containing the trained model. To avoid having to train a model from scratch, there is a already trained model for spinal cord segmentation in the folder named trained_model, in the downloaded dataset. Modify the path so it points to the location of the trained model."path_output": "<PATH_TO_DATASET>/data_example_spinegeneric/trained_model"Note that you can also pass this argument via CLI (see Usage)
ivadomed -c path/to/config --path-output path/to/output/directory
command: Action to perform. Here, we want to do some inference using the previously trained model, so we set the field as follows:"command": "test"Note that you can also pass this argument via CLI (see Usage)
ivadomed --test -c path/to/config
uncertainty: Type of uncertainty to estimate. Available choices areepistemicandaleatoric. Note that both can betrue. More details on the implementation are available in Technical features.n_itcontrols the number of Monte Carlo iterations that are performed to estimate the uncertainty. Set it to a non-zero positive integer for this tutorial (e.g.20)."uncertainty": { "epistemic": true, "aleatoric": true, "n_it": 20 }
transformation: Data augmentation transformation. If you have selected the aleatoric uncertainty, the data augmentation that will be performed is the same as the one performed for the training. Note that only transformations for which aundo_transform(i.e. inverse transformation) is available will be performed since these inverse transformations are required to reconstruct the predicted volume.
Run uncertainty estimation
Once the configuration file has been modified, run the inference with the following command:
ivadomed --test -c config.json --path-data <PATH_TO_DATASET>/data_example_spinegeneric --path-output <PATH_TO_DATASET>/data_example_spinegeneric/trained_model
Here, we want to do some inference using the previously trained model, so we set the command flag as follows:
--test
--path-data: Location of the directory containing the dataset.--path-data <PATH_TO_DATASET>/data_example_spinegeneric
--path-output: Folder name that will contain the output files (e.g., trained model, predictions, results). For the purpose of this particular tutorial, since we do not train the model from scratch, we set the output path to point to a folder containing the pre-trained model for spinal cord segmentation that comes with the dataset. Hence, after running this tutorial, the corresponding output files can be found inside the trained_model folder.--path-output <PATH_TO_DATASET>/data_example_spinegeneric/trained_modelIf you set the
command,path_output, andpath_dataarguments in your config file, you do not need to pass the CLI flags:ivadomed -c config.jsonIf aleatoric uncertainty was selected, then data augmentation operations will be performed at testing time, as indicated in the terminal output (see below). Note that
ElasticTransformhas been deactivated because noundo_transformfunction is available for it.Selected transformations for the ['testing'] dataset: Resample: {'hspace': 0.75, 'wspace': 0.75, 'dspace': 1} CenterCrop: {'size': [128, 128]} RandomAffine: {'degrees': 5, 'scale': [0.1, 0.1], 'translate': [0.03, 0.03], 'applied_to': ['im', 'gt']} ElasticTransform: {'alpha_range': [28.0, 30.0], 'sigma_range': [3.5, 4.5], 'p': 0.1, 'applied_to': ['im', 'gt']} NumpyToTensor: {} NormalizeInstance: {'applied_to': ['im']} ElasticTransform transform not included since no undo_transform available for it.… otherwise, only preprocessing and data normalization are performed, see below:
Selected transformations for the ['testing'] dataset: Resample: {'hspace': 0.75, 'wspace': 0.75, 'dspace': 1} CenterCrop: {'size': [128, 128]} NumpyToTensor: {} NormalizeInstance: {'applied_to': ['im']}For each Monte Carlo iteration, each testing image is segmented using the trained model and saved under
pred_masks, with the iteration number as suffix (e.g.sub-001_pred_00.nii.gz…sub-001_pred_19.nii.gz).Computing model uncertainty over 20 iterations. Inference - Iteration 0: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:11<00:00, 2.27s/it] Inference - Iteration 1: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.81s/it] Inference - Iteration 2: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.96s/it] Inference - Iteration 3: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:08<00:00, 1.66s/it] Inference - Iteration 4: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:08<00:00, 1.69s/it] Inference - Iteration 5: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.92s/it] Inference - Iteration 6: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:08<00:00, 1.74s/it] Inference - Iteration 7: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:08<00:00, 1.74s/it] Inference - Iteration 8: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.83s/it] Inference - Iteration 9: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [00:07<00:00, 1.59s/it] Inference - Iteration 10: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.85s/it] Inference - Iteration 11: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.85s/it] Inference - Iteration 12: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.92s/it] Inference - Iteration 13: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.83s/it] Inference - Iteration 14: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.84s/it] Inference - Iteration 15: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.87s/it] Inference - Iteration 16: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.81s/it] Inference - Iteration 17: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.95s/it] Inference - Iteration 18: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:09<00:00, 1.82s/it] Inference - Iteration 19: 100%|██████████████████████████████████████████████████████████████████████████████████| 5/5 [00:08<00:00, 1.71s/it]The Monte Carlo samples are then used to compute uncertainty measures for each image. The results are saved under
pred_masks.Uncertainty Computation: 100%|███████████████████████████████████████████████████████████████████████████████████| 5/5 [01:31<00:00, 18.28s/it]Six files are generated during this process for each testing image:
*_soft.nii.gz: Soft segmentation (i.e. values between 0 and 1) which is generated by averaging the Monte Carlo samples.
*_pred.nii.gz: Binary segmentation obtained by thresholding*_soft.nii.gzwith1 / (Number of Monte Carlo iterations).
*_unc-vox.nii.gz: Voxel-wise measure of uncertainty derived from the entropy of the Monte Carlo samples. The higher a given voxel value is, the more uncertain is the prediction for this voxel.
*_unc-avgUnc.nii.gz: Structure-wise measure of uncertainty derived from the mean value of*_unc-vox.nii.gzwithin a given connected object (e.g. a lesion, grey matter).
*_unc-cv.nii.gz: Structure-wise measure of uncertainty derived from the coefficient of variation of the volume of a given connected object across the Monte Carlo samples. The higher a given voxel value is, the more uncertain is the prediction for this voxel.
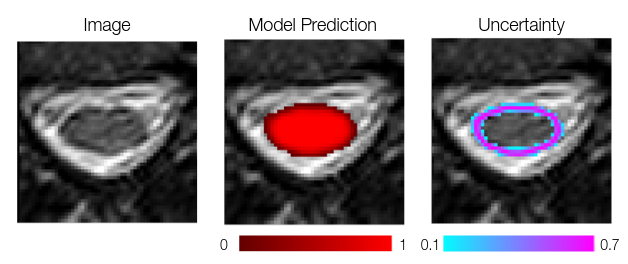
*_unc-iou.nii.gz: Structure-wise measure of uncertainty derived from the Intersection-over-Union of the predictions of a given connected object across the Monte Carlo samples. The lower a given voxel value is, the more uncertain is the prediction for this voxel.These files can further be used for post-processing to refine the segmentation. For example, the voxels depicted in pink are more uncertain than the ones in blue (left image): we might want to refine the model prediction by removing from the foreground class the voxels with low uncertainty (blue, left image) AND low prediction value (dark red, middle image).