API Reference
This document is for developers of ivadomed, it contains the API functions.
Loader API
loader.film
- normalize_metadata(ds_in: BidsDataset | Bids3DDataset | MRI2DSegmentationDataset, clustering_models: dict, debugging: bool, metadata_type: str, train_set: bool = False) list, OneHotEncoder | list[source]
Categorize each metadata value using a KDE clustering method, then apply a one-hot-encoding.
- Parameters:
ds_in (BidsDataset) – Dataset BidsDataset, Bids3D, MRI2D with metadata.
clustering_models (dict) – Pre-trained clustering model that has been trained on metadata of the training set.
debugging (bool) – If True, extended verbosity and intermediate outputs.
metadata_type (str) – Choice between ‘mri_params’, ‘constrasts’ or the name of a column from the participants.tsv file.
train_set (bool) – Indicates if the input dataset is the training dataset (True) or the validation or testing dataset (False).
- Returns:
- Dataset with normalized metadata. If train_set is True, then the one-hot-encoder model is also
returned.
- Return type:
BidsDataset
- class Kde_model[source]
Bases:
objectKernel Density Estimation.
Apply this clustering method to metadata values, using (sklearn implementation.)
- Attributes:
kde (sklearn.neighbors.KernelDensity)
minima (float) – Local minima.
- clustering_fit(dataset: list, key_lst: List[str]) dict[source]
This function creates clustering models for each metadata type, using Kernel Density Estimation algorithm.
- Parameters:
dataset (list) – data
key_lst (list of str) – names of metadata to cluster
- Returns:
Clustering model for each metadata type in a dictionary where the keys are the metadata names.
- Return type:
dict
- check_isMRIparam(mri_param_type: str, mri_param: dict, subject: str, metadata: dict) bool[source]
Check if a given metadata belongs to the MRI parameters.
- Parameters:
mri_param_type (str) – Metadata type name.
mri_param (dict) – List of MRI params names.
subject (str) – Current subject name.
metadata (dict) – Metadata.
- Returns:
True if mri_param_type is part of mri_param.
- Return type:
bool
- get_film_metadata_models(ds_train: MRI2DSegmentationDataset, metadata_type: str, debugging: bool = False)[source]
Get FiLM models.
This function pulls the clustering and one-hot encoder models that are used by FiLMedUnet. It also calls the normalization of metadata.
- Parameters:
ds_train (MRI2DSegmentationDataset) – training dataset
metadata_type (string) – eg mri_params, contrasts
debugging (bool) –
- Returns:
dataset, one-hot encoder and KDE model
- Return type:
MRI2DSegmentationDataset, OneHotEncoder, KernelDensity
- store_film_params(gammas: dict, betas: dict, metadata_values: list, metadata: list, model: nn.Module, film_layers: list, depth: int, film_metadata: str)[source]
Store FiLM params.
- Parameters:
gammas (dict) –
betas (dict) –
metadata_values (list) – list of the batch sample’s metadata values (e.g., T2w, astrocytoma)
metadata (list) –
model (nn.Module) –
film_layers (list) –
depth (int) –
film_metadata (str) – Metadata of interest used to modulate the network (e.g., contrast, tumor_type).
- Returns:
gammas, betas, metadata_values
- Return type:
dict, dict, list
- save_film_params(gammas: dict, betas: dict, metadata_values: list, depth: int, ofolder: str) None[source]
Save FiLM params as npy files.
These parameters can be further used for visualisation purposes. They are saved in the ofolder with .npy format.
- Parameters:
gammas (dict) –
betas (dict) –
metadata_values (list) – list of the batch sample’s metadata values (eg T2w, T1w, if metadata type used is
contrast) –
depth (int) –
ofolder (str) –
loader.loader
- load_dataset(bids_df: BidsDataframe, data_list: List[str], transforms_params: dict, model_params: dict, target_suffix: List[str], roi_params: dict, contrast_params: dict, slice_filter_params: dict, patch_filter_params: dict, slice_axis: str, multichannel: bool, dataset_type: str = 'training', requires_undo: bool = False, metadata_type: str | None = None, object_detection_params: dict | None = None, soft_gt: bool = False, device: device | None = None, cuda_available: bool | None = None, is_input_dropout: bool = False, **kwargs) Bids3DDataset[source]
Get loader appropriate loader according to model type. Available loaders are Bids3DDataset for 3D data, BidsDataset for 2D data and HDF5Dataset for HeMIS.
- Parameters:
bids_df (BidsDataframe) – Object containing dataframe with all BIDS image files and their metadata.
data_list (list) – Subject names list.
transforms_params (dict) – Dictionary containing transformations for “training”, “validation”, “testing” (keys), eg output of imed_transforms.get_subdatasets_transforms.
model_params (dict) – Dictionary containing model parameters.
target_suffix (list of str) – List of suffixes for target masks.
roi_params (dict) – Contains ROI related parameters.
contrast_params (dict) – Contains image contrasts related parameters.
slice_filter_params (dict) – Contains slice_filter_params, see Configuration File for more details.
patch_filter_params (dict) – Contains patch_filter_params, see Configuration File for more details.
slice_axis (string) – Choice between “axial”, “sagittal”, “coronal” ; controls the axis used to extract the 2D data from 3D NifTI files. 2D PNG/TIF/JPG files use default “axial.
multichannel (bool) – If True, the input contrasts are combined as input channels for the model. Otherwise, each contrast is processed individually (ie different sample / tensor).
metadata_type (str) – Choice between None, “mri_params”, “contrasts”.
dataset_type (str) – Choice between “training”, “validation” or “testing”.
requires_undo (bool) – If True, the transformations without undo_transform will be discarded.
object_detection_params (dict) – Object dection parameters.
soft_gt (bool) – If True, ground truths are not binarized before being fed to the network. Otherwise, ground
thresholded (truths are) –
device (torch.device) – Device to use for the model training.
cuda_available (bool) – If True, cuda is available.
is_input_dropout (bool) – Return input with missing modalities.
- Returns:
BidsDataset
Note: For more details on the parameters transform_params, target_suffix, roi_params, contrast_params, slice_filter_params, patch_filter_params and object_detection_params see Configuration File.
loader.utils
- split_dataset(df: pd.DataFrame, split_method: str, data_testing: dict, random_seed: int, train_frac: float = 0.8, test_frac: float = 0.1)[source]
Splits dataset into training, validation and testing sets by applying train, test and validation fractions according to the split_method. The “data_testing” parameter can be used to specify the data_type and data_value to include in the testing set, the dataset is then split as not to mix the data_testing between the training/validation set and the testing set.
- Parameters:
df (pd.DataFrame) – Dataframe containing all BIDS image files indexed and their metadata.
split_method (str) – Used to specify on which metadata to split the dataset (eg. “participant_id”, “sample_id”, etc.)
data_testing (dict) – Used to specify data_type and data_value to include in the testing set.
random_seed (int) – Random seed to ensure reproducible splits.
train_frac (float) – Between 0 and 1. Represents the train set proportion.
test_frac (float) – Between 0 and 1. Represents the test set proportion.
- Returns:
Train, validation and test filenames lists.
- Return type:
list, list, list
- get_new_subject_file_split(df: pd.DataFrame, split_method: str, data_testing: dict, random_seed: int, train_frac: float, test_frac: float, path_output: str, balance: str, subject_selection: dict = None)[source]
Randomly split dataset between training / validation / testing.
Randomly split dataset between training / validation / testing and save it in path_output + “/split_datasets.joblib”.
- Parameters:
df (pd.DataFrame) – Dataframe containing all BIDS image files indexed and their metadata.
split_method (str) – Used to specify on which metadata to split the dataset (eg. “participant_id”, “sample_id”, etc.)
data_testing (dict) – Used to specify the data_type and data_value to include in the testing set.
random_seed (int) – Random seed.
train_frac (float) – Training dataset proportion, between 0 and 1.
test_frac (float) – Testing dataset proportionm between 0 and 1.
path_output (str) – Output folder.
balance (str) – Metadata contained in “participants.tsv” file with categorical values. Each category will be
training (evenly distributed in the) –
datasets. (validation and testing) –
subject_selection (dict) – Used to specify a custom subject selection from a dataset.
- Returns:
Training, validation and testing filenames lists.
- Return type:
list, list list
- get_subdatasets_subject_files_list(split_params: dict, df: pd.DataFrame, path_output: str, subject_selection: dict = None)[source]
Get lists of subject filenames for each sub-dataset between training / validation / testing.
- Parameters:
split_params (dict) – Split parameters, see Configuration File for more details.
df (pd.DataFrame) – Dataframe containing all BIDS image files indexed and their metadata.
path_output (str) – Output folder.
subject_selection (dict) – Used to specify a custom subject selection from a dataset.
- Returns:
Training, validation and testing filenames lists.
- Return type:
list, list list
- imed_collate(batch: dict) dict | list | str | Tensor[source]
Collates data to create batches
- Parameters:
batch (dict) – Contains input and gt data with their corresponding metadata.
- Returns:
Collated data.
- Return type:
list or dict or str or tensor
- filter_roi(roi_data: ndarray, nb_nonzero_thr: int) bool[source]
Filter slices from dataset using ROI data.
This function filters slices (roi_data) where the number of non-zero voxels within the ROI slice (e.g. centerline, SC segmentation) is inferior or equal to a given threshold (nb_nonzero_thr).
- Parameters:
roi_data (nd.array) – ROI slice.
nb_nonzero_thr (int) – Threshold.
- Returns:
True if the slice needs to be filtered, False otherwise.
- Return type:
bool
- orient_img_hwd(data: ndarray, slice_axis: int) ndarray[source]
Orient a given RAS image to height, width, depth according to slice axis.
- Parameters:
data (ndarray) – RAS oriented data.
slice_axis (int) – Indicates the axis used for the 2D slice extraction: Sagittal: 0, Coronal: 1, Axial: 2.
- Returns:
Array oriented with the following dimensions: (height, width, depth).
- Return type:
ndarray
- orient_img_ras(data: ndarray, slice_axis: int) ndarray[source]
Orient a given array with dimensions (height, width, depth) to RAS orientation.
- Parameters:
data (ndarray) – Data with following dimensions (Height, Width, Depth).
slice_axis (int) – Indicates the axis used for the 2D slice extraction: Sagittal: 0, Coronal: 1, Axial: 2.
- Returns:
Array oriented in RAS.
- Return type:
ndarray
- orient_shapes_hwd(data: list | tuple, slice_axis: int) ndarray[source]
Swap dimensions according to match the height, width, depth orientation.
- Parameters:
data (list or tuple) – Shape or numbers associated with each image dimension (e.g. image resolution).
slice_axis (int) – Indicates the axis used for the 2D slice extraction: Sagittal: 0, Coronal: 1, Axial: 2.
- Returns:
Reoriented vector.
- Return type:
ndarray
- update_metadata(metadata_src_lst: list, metadata_dest_lst: list) list[source]
Update metadata keys with a reference metadata.
A given list of metadata keys will be changed and given the values of the reference metadata.
- Parameters:
metadata_src_lst (list) – List of source metadata used as reference for the destination metadata.
metadata_dest_lst (list) – List of metadate that needs to be updated.
- Returns:
updated metadata list.
- Return type:
list
- reorient_image(arr: np.ndarray, slice_axis: int, nib_ref: nib, nib_ref_canonical: nib) nd.ndarray[source]
Reorient an image to match a reference image orientation.
It reorients a array to a given orientation and convert it to a nibabel object using the reference nibabel header.
- Parameters:
arr (ndarray) – Input array, array to re orient.
slice_axis (int) – Indicates the axis used for the 2D slice extraction: Sagittal: 0, Coronal: 1, Axial: 2.
nib_ref (nibabel) – Reference nibabel object, whose header is used.
nib_ref_canonical (nibabel) – nib_ref that has been reoriented to canonical orientation (RAS).
- get_file_extension(filename: str) str | None[source]
Get file extension if it is supported :param filename: Path of the file. :type filename: str
- Returns:
File extension
- Return type:
str
- update_filename_to_nifti(filename: str) str[source]
Update filename extension to ‘nii.gz’ if not a NifTI file.
This function is used to help make non-NifTI files (e.g. PNG/TIF/JPG) compatible with NifTI-only pipelines. The expectation is that a NifTI version of the file has been created alongside the original file, which allows the extension to be cleanly swapped for a .nii.gz extension.
- Parameters:
filename (str) – Path of original file.
- Returns:
Path of the corresponding NifTI file.
- Return type:
str
- dropout_input(seg_pair: dict) dict[source]
Applies input-level dropout: zero to all channels minus one will be randomly set to zeros. This function verifies if some channels are already empty. Always at least one input channel will be kept.
- Parameters:
seg_pair (dict) – Batch containing torch tensors (input and gt) and metadata.
- Returns:
Batch containing torch tensors (input and gt) and metadata with channel(s) dropped.
- Return type:
seg_pair (dict)
- create_temp_directory() str[source]
Creates a temporary directory and returns its path. This temporary directory is only deleted when explicitly requested.
- Returns:
Path of the temporary directory.
- Return type:
str
- get_obj_size(obj) int[source]
Returns the size of an object in bytes. Used to gauge whether storing object in memory vs write to disk.
Source: https://stackoverflow.com/a/53705610
- Parameters:
obj –
Returns:
Object Detection API
object_detection.utils
- get_bounding_boxes(mask)[source]
Generates a 3D bounding box around a given mask. :param mask: Mask of the ROI. :type mask: Numpy array
- Returns:
Bounding box coordinate (x_min, x_max, y_min, y_max, z_min, z_max).
- Return type:
list
- adjust_bb_size(bounding_box, factor, resample=False)[source]
Modifies the bounding box dimensions according to a given factor.
- Parameters:
bounding_box (list or tuple) – Coordinates of bounding box (x_min, x_max, y_min, y_max, z_min, z_max).
factor (list or tuple) – Multiplicative factor for each dimension (list or tuple of length 3).
resample (bool) – Boolean indicating if this resize is for resampling.
- Returns:
New coordinates (x_min, x_max, y_min, y_max, z_min, z_max).
- Return type:
list
- resize_to_multiple(shape, multiple, length)[source]
Modify a given shape so each dimension is a multiple of a given number. This is used to avoid dimension mismatch with patch training. The return shape is always larger then the initial shape (no cropping).
- Parameters:
shape (tuple or list) – Initial shape to be modified.
multiple (tuple or list) – Multiple for each dimension.
length (tuple or list) – Patch length.
- Returns:
New dimensions.
- Return type:
list
- generate_bounding_box_file(subject_path_list, model_path, path_output, gpu_id=0, slice_axis=0, contrast_lst=None, keep_largest_only=True, safety_factor=None)[source]
Creates json file containing the bounding box dimension for each images. The file has the following format: {“path/to/img.nii.gz”: [[x1_min, x1_max, y1_min, y1_max, z1_min, z1_max], [x2_min, x2_max, y2_min, y2_max, z2_min, z2_max]]} where each list represents the coordinates of an object on the image (2 instance of a given object in this example).
- Parameters:
subject_path_list (list) – List of all subjects in all the BIDS directories.
model_path (string) – Path to object detection model.
path_output (string) – Output path.
gpu_id (int) – If available, GPU number.
slice_axis (int) – Slice axis (0: sagittal, 1: coronal, 2: axial).
contrast_lst (list) – Contrasts.
keep_largest_only (bool) – Boolean representing if only the largest object of the prediction is kept.
safety_factor (list or tuple) – Factors to multiply each dimension of the bounding box.
- Returns:
Dictionary containing bounding boxes related to their image.
- Return type:
dict
- resample_bounding_box(metadata, transform)[source]
Resample bounding box.
- Parameters:
metadata (dict) – Dictionary containing the metadata to be modified with the resampled coordinates.
transform (Compose) – Transformations possibly containing the resample params.
- adjust_transforms(transforms, seg_pair, length=None, stride=None)[source]
This function adapts the transforms by adding the BoundingBoxCrop transform according the specific parameters of an image. The dimensions of the crop are also adapted to fit the length and stride parameters if the 3D loader is used.
- adjust_undo_transforms(transforms, seg_pair, index=0)[source]
This function adapts the undo transforms by adding the BoundingBoxCrop to undo transform according the specific parameters of an image.
- Parameters:
transforms (Compose) – Transforms.
seg_pair (dict) – Segmentation pair (input, gt and metadata).
index (int) – Batch index of the seg_pair.
- load_bounding_boxes(object_detection_params, subject_path_list, slice_axis, constrast_lst)[source]
Verifies if bounding_box.json exists in the output path, if so loads the data, else generates the file if a valid detection model path exists.
- Parameters:
object_detection_params (dict) – Object detection parameters.
subject_path_list (list) – List of all subjects in all the BIDS directories.
slice_axis (int) – Slice axis (0: sagittal, 1: coronal, 2: axial).
constrast_lst (list) – Contrasts.
- Returns:
bounding boxes for every subject in BIDS directory
- Return type:
dict
- verify_metadata(metadata, has_bounding_box)[source]
Validates across all metadata that the ‘bounding_box’ param is present.
- Parameters:
metadata (dict) – Image metadata.
has_bounding_box (bool) – If ‘bounding_box’ is present across all metadata, True, else False.
- Returns:
Boolean indicating if ‘bounding_box’ is present across all metadata.
- Return type:
bool
- bounding_box_prior(fname_mask, metadata, slice_axis, safety_factor=None)[source]
Computes prior steps to a model requiring bounding box crop. This includes loading a mask of the ROI, orienting the given mask into the following dimensions: (height, width, depth), extracting the bounding boxes and storing the information in the metadata.
- Parameters:
fname_mask (str) – Filename containing the mask of the ROI
metadata (dict) – Dictionary containing the image metadata
slice_axis (int) – Slice axis (0: sagittal, 1: coronal, 2: axial)
safety_factor (list or tuple) – Factors to multiply each dimension of the bounding box.
Evaluation API
- evaluate(bids_df, path_output, target_suffix, eval_params)[source]
Evaluate predictions from inference step.
- Parameters:
bids_df (BidsDataframe) – Object containing dataframe with all BIDS image files and their metadata.
path_output (str) – Folder where the output folder “results_eval” is be created.
target_suffix (list) – List of suffixes that indicates the target mask(s).
eval_params (dict) – Evaluation parameters.
- Returns:
results for each image.
- Return type:
pd.Dataframe
- class Evaluation3DMetrics(data_pred, data_gt, dim_lst, params=None)[source]
Bases:
objectComputes 3D evaluation metrics.
- Parameters:
data_pred (ndarray) – Network prediction mask.
data_gt (ndarray) – Ground-truth mask.
dim_lst (list) – Resolution (mm) along each dimension.
params (dict) – Evaluation parameters.
- Attributes:
data_pred (ndarray) – Network prediction mask.
data_gt (ndarray) – Ground-truth mask.
n_classes (int) – Number of classes.
px (float) – Resolution (mm) along the first axis.
py (float) – Resolution (mm) along the second axis.
pz (float) – Resolution (mm) along the third axis.
bin_struct (ndarray) – Binary structure.
size_min (int) – Minimum size of objects. Objects that are smaller than this limit can be removed if “removeSmall” is in params.
object_detection_metrics (bool) – Indicate if object detection metrics (lesions true positive and false detection rates) are computed or not.
overlap_vox (int) – A prediction and ground-truth are considered as overlapping if they overlap for at least this amount of voxels.
overlap_ratio (float) – A prediction and ground-truth are considered as overlapping if they overlap for at least this portion of their volumes.
data_pred_label (ndarray) – Network prediction mask that is labeled, ie each object is filled with a different value.
data_gt_label (ndarray) – Ground-truth mask that is labeled, ie each object is filled with a different value.
n_pred (int) – Number of objects in the network prediction mask.
n_gt (int) – Number of objects in the ground-truth mask.
data_painted (ndarray) – Mask where each predicted object is labeled depending on whether it is a TP or FP.
- label_per_size(data)[source]
Get data with labels corresponding to label size.
- Parameters:
data (ndarray) – Input data.
- Returns:
ndarray
- get_rvd()[source]
Relative volume difference.
The volume is here defined by the physical volume, in mm3, of the non-zero voxels of a given mask. Relative volume difference equals the difference between the ground-truth and prediction volumes, divided by the ground-truth volume. Optimal value is zero. Negative value indicates over-segmentation, while positive value indicates under-segmentation.
- get_avd()[source]
Absolute volume difference.
The volume is here defined by the physical volume, in mm3, of the non-zero voxels of a given mask. Absolute volume difference equals the absolute value of the Relative Volume Difference. Optimal value is zero.
- get_ltpr(label_size=None, class_idx=0)[source]
Lesion True Positive Rate / Recall / Sensitivity.
- Parameters:
label_size (int) – Size of label.
class_idx (int) – Label index. If monolabel 0, else ranges from 0 to number of output channels - 1.
Note: computed only if n_obj >= 1 and “object_detection_metrics” evaluation parameter is True.
- get_lfdr(label_size=None, class_idx=0)[source]
Lesion False Detection Rate / 1 - Precision.
- Parameters:
label_size (int) – Size of label.
class_idx (int) – Label index. If monolabel 0, else ranges from 0 to number of output channels - 1.
Note: computed only if n_obj >= 1 and “object_detection_metrics” evaluation parameter is True.
Losses API
- class MultiClassDiceLoss(classes_of_interest=None)[source]
Bases:
ModuleMulti-class Dice Loss.
Inspired from https://arxiv.org/pdf/1802.10508.
- Parameters:
classes_of_interest (list) – List containing the index of a class which its dice will be added to the loss. If is None all classes are considered.
- Attributes:
classes_of_interest (list) – List containing the index of a class which its dice will be added to the loss. If is None all classes are considered.
dice_loss (DiceLoss) – Class computing the Dice loss.
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(classes_of_interest=None)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(prediction, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class DiceLoss(smooth=1.0)[source]
Bases:
ModuleDiceLoss.
See also
Milletari, Fausto, Nassir Navab, and Seyed-Ahmad Ahmadi. “V-net: Fully convolutional neural networks for volumetric medical image segmentation.” 2016 fourth international conference on 3D vision (3DV). IEEE, 2016.
- Parameters:
smooth (float) – Value to avoid division by zero when images and predictions are empty.
- Attributes:
smooth (float) – Value to avoid division by zero when images and predictions are empty.
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(smooth=1.0)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(prediction, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class BinaryCrossEntropyLoss[source]
Bases:
Module- Attributes:
loss_fct (BCELoss) – Binary cross entropy loss function from torch library.
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(prediction, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class FocalLoss(gamma=2, alpha=0.25, eps=1e-07)[source]
Bases:
ModuleFocalLoss.
See also
Lin, Tsung-Yi, et al. “Focal loss for dense object detection.” Proceedings of the IEEE international conference on computer vision. 2017.
- Parameters:
gamma (float) – Value from 0 to 5, Control between easy background and hard ROI training examples. If set to 0, equivalent to cross-entropy.
alpha (float) – Value from 0 to 1, usually corresponding to the inverse of class frequency to address class imbalance.
eps (float) – Epsilon to avoid division by zero.
- Attributes:
gamma (float) – Value from 0 to 5, Control between easy background and hard ROI training examples. If set to 0, equivalent to cross-entropy.
alpha (float) – Value from 0 to 1, usually corresponding to the inverse of class frequency to address class imbalance.
eps (float) – Epsilon to avoid division by zero.
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(gamma=2, alpha=0.25, eps=1e-07)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(input, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class FocalDiceLoss(beta=1, gamma=2, alpha=0.25)[source]
Bases:
ModuleFocalDiceLoss.
See also
Wong, Ken CL, et al. “3D segmentation with exponential logarithmic loss for highly unbalanced object sizes.” International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2018.
- Parameters:
beta (float) – Value from 0 to 1, indicating the weight of the dice loss.
gamma (float) – Value from 0 to 5, Control between easy background and hard ROI training examples. If set to 0, equivalent to cross-entropy.
alpha (float) – Value from 0 to 1, usually corresponding to the inverse of class frequency to address class imbalance.
- Attributes:
beta (float) – Value from 0 to 1, indicating the weight of the dice loss.
gamma (float) – Value from 0 to 5, Control between easy background and hard ROI training examples. If set to 0, equivalent to cross-entropy.
alpha (float) – Value from 0 to 1, usually corresponding to the inverse of class frequency to address class imbalance.
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(beta=1, gamma=2, alpha=0.25)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(input, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class GeneralizedDiceLoss(epsilon=1e-05, include_background=True)[source]
Bases:
ModuleGeneralizedDiceLoss.
See also
Sudre, Carole H., et al. “Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations.” Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, Cham, 2017. 240-248.
- Parameters:
epsilon (float) – Epsilon to avoid division by zero.
include_background (bool) – If True, then an extra channel is added, which represents the background class.
- Attributes:
epsilon (float) – Epsilon to avoid division by zero.
include_background (bool) – If True, then an extra channel is added, which represents the background class.
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(epsilon=1e-05, include_background=True)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(input, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class TverskyLoss(alpha=0.7, beta=0.3, smooth=1.0)[source]
Bases:
ModuleTversky Loss.
See also
Salehi, Seyed Sadegh Mohseni, Deniz Erdogmus, and Ali Gholipour. “Tversky loss function for image segmentation using 3D fully convolutional deep networks.” International Workshop on Machine Learning in Medical Imaging. Springer, Cham, 2017.
- Parameters:
alpha (float) – Weight of false positive voxels.
beta (float) – Weight of false negative voxels.
smooth (float) – Epsilon to avoid division by zero, when both Numerator and Denominator of Tversky are zeros.
- Attributes:
alpha (float) – Weight of false positive voxels.
beta (float) – Weight of false negative voxels.
smooth (float) – Epsilon to avoid division by zero, when both Numerator and Denominator of Tversky are zeros.
Notes
setting alpha=beta=0.5: Equivalent to DiceLoss.
default parameters were suggested by https://arxiv.org/pdf/1706.05721.pdf .
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(alpha=0.7, beta=0.3, smooth=1.0)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- tversky_index(y_pred, y_true)[source]
Compute Tversky index.
- Parameters:
y_pred (torch Tensor) – Prediction.
y_true (torch Tensor) – Target.
- Returns:
Tversky index.
- Return type:
float
- forward(input, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class FocalTverskyLoss(alpha=0.7, beta=0.3, gamma=1.33, smooth=1.0)[source]
Bases:
TverskyLossFocal Tversky Loss.
See also
Abraham, Nabila, and Naimul Mefraz Khan. “A novel focal tversky loss function with improved attention u-net for lesion segmentation.” 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). IEEE, 2019.
- Parameters:
alpha (float) – Weight of false positive voxels.
beta (float) – Weight of false negative voxels.
gamma (float) – Typically between 1 and 3. Control between easy background and hard ROI training examples.
smooth (float) – Epsilon to avoid division by zero, when both Numerator and Denominator of Tversky are zeros.
- Attributes:
gamma (float) – Typically between 1 and 3. Control between easy background and hard ROI training examples.
Notes
setting alpha=beta=0.5 and gamma=1: Equivalent to DiceLoss.
default parameters were suggested by https://arxiv.org/pdf/1810.07842.pdf .
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(alpha=0.7, beta=0.3, gamma=1.33, smooth=1.0)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(input, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class L2loss[source]
Bases:
ModuleEuclidean loss also known as L2 loss. Compute the sum of the squared difference between the two images.
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(input, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class AdapWingLoss(theta=0.5, alpha=2.1, omega=14, epsilon=1)[source]
Bases:
ModuleAdaptive Wing loss Used for heatmap ground truth.
See also
Wang, Xinyao, Liefeng Bo, and Li Fuxin. “Adaptive wing loss for robust face alignment via heatmap regression.” Proceedings of the IEEE International Conference on Computer Vision. 2019.
- Parameters:
theta (float) – Threshold between linear and non linear loss.
alpha (float) – Used to adapt loss shape to input shape and make loss smooth at 0 (background).
properties. (It needs to be slightly above 2 to maintain ideal) –
omega (float) – Multiplicating factor for non linear part of the loss.
epsilon (float) – factor to avoid gradient explosion. It must not be too small
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(theta=0.5, alpha=2.1, omega=14, epsilon=1)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(input, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
- class LossCombination(losses_list, params_list=None)[source]
Bases:
ModuleLoss that sums other implemented losses.
- Parameters:
losses_list (list) – list of losses that will be summed. Elements should be string.
params_list (list) – list of params for the losses, contain None or dictionnary definition of params for the loss
used. (at same index. If no params list is given all default parameter will be) –
(e.g. – params_list = [None,{“param1:0.5”}])
["L2loss" (losses_list =) – params_list = [None,{“param1:0.5”}])
"DiceLoss"] – params_list = [None,{“param1:0.5”}])
- Returns:
sum of losses computed on (input,target) with the params
- Return type:
tensor
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(losses_list, params_list=None)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(input, target)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
Main API
- run_command(context, n_gif=0, thr_increment=None, resume_training=False, no_patch=False, overlap_2d=None)[source]
Run main command.
This function is central in the ivadomed project as training / testing / evaluation commands are run via this function. All the process parameters are defined in the config.
- Parameters:
context (dict) – Dictionary containing all parameters that are needed for a given process. See Configuration File for more details.
n_gif (int) – Generates a GIF during training if larger than zero, one frame per epoch for a given slice. The parameter indicates the number of 2D slices used to generate GIFs, one GIF per slice. A GIF shows predictions of a given slice from the validation sub-dataset. They are saved within the output path.
thr_increment (float) – A threshold analysis is performed at the end of the training using the trained model and the training + validation sub-dataset to find the optimal binarization threshold. The specified value indicates the increment between 0 and 1 used during the ROC analysis (e.g. 0.1).
resume_training (bool) – Load a saved model (“checkpoint.pth.tar” in the output directory specified with flag “–path-output” or via the config file “output_path”. This training state is saved everytime a new best model is saved in the log argument) for resume training directory.
no_patch (bool) – If True, 2D patches are not used while segmenting with models trained with patches (command “–segment” only). Default: False (i.e. segment with patches). The “no_patch” option supersedes the “overlap_2D” option.
overlap_2d (list of int) – Custom overlap for 2D patches while segmenting (command “–segment” only). Default model overlap is used otherwise.
- Returns:
If “train” command: Returns floats: best loss score for both training and validation.
If “test” command: Returns a pandas Dataframe: of metrics computed for each subject of the testing sub-dataset and return the prediction metrics before evaluation.
If “segment” command: No return value.
- Return type:
float or pandas.DataFrame or None
Metrics API
- class MetricManager(metric_fns)[source]
Bases:
objectComputes specified metrics and stores them in a dictionary.
- Parameters:
metric_fns (list) – List of metric functions.
- Attributes:
metric_fns (list) – List of metric functions.
result_dict (dict) – Dictionary storing metrics.
num_samples (int) – Number of samples.
- numeric_score(prediction, groundtruth)[source]
Computation of statistical numerical scores:
FP = Soft False Positives
FN = Soft False Negatives
TP = Soft True Positives
TN = Soft True Negatives
- Robust to hard or soft input masks. For example::
prediction=np.asarray([0, 0.5, 1]) groundtruth=np.asarray([0, 1, 1]) Leads to FP = 1.5
Note: It assumes input values are between 0 and 1.
- Parameters:
prediction (ndarray) – Binary prediction.
groundtruth (ndarray) – Binary groundtruth.
- Returns:
FP, FN, TP, TN
- Return type:
float, float, float, float
- dice_score(im1, im2, empty_score=nan)[source]
Computes the Dice coefficient between im1 and im2.
Compute a soft Dice coefficient between im1 and im2, ie equals twice the sum of the two masks product, divided by the sum of each mask sum. If both images are empty, then it returns empty_score.
- Parameters:
im1 (ndarray) – First array.
im2 (ndarray) – Second array.
empty_score (float) – Returned value if both input array are empty.
- Returns:
Dice coefficient.
- Return type:
float
- mse(im1, im2)[source]
Compute the Mean Squared Error.
Compute the Mean Squared Error between the two images, i.e. sum of the squared difference.
- Parameters:
im1 (ndarray) – First array.
im2 (ndarray) – Second array.
- Returns:
Mean Squared Error.
- Return type:
float
- hausdorff_score(prediction, groundtruth)[source]
Compute the directed Hausdorff distance between two N-D arrays.
- Parameters:
prediction (ndarray) – First array.
groundtruth (ndarray) – Second array.
- Returns:
Hausdorff distance.
- Return type:
float
- precision_score(prediction, groundtruth, err_value=0.0)[source]
Positive predictive value (PPV).
Precision equals the number of true positive voxels divided by the sum of true and false positive voxels. True and false positives are computed on soft masks, see
"numeric_score".- Parameters:
prediction (ndarray) – First array.
groundtruth (ndarray) – Second array.
err_value (float) – Value returned in case of error.
- Returns:
Precision score.
- Return type:
float
- recall_score(prediction, groundtruth, err_value=0.0)[source]
True positive rate (TPR).
Recall equals the number of true positive voxels divided by the sum of true positive and false negative voxels. True positive and false negative values are computed on soft masks, see
"numeric_score".- Parameters:
prediction (ndarray) – First array.
groundtruth (ndarray) – Second array.
err_value (float) – Value returned in case of error.
- Returns:
Recall score.
- Return type:
float
- specificity_score(prediction, groundtruth, err_value=0.0)[source]
True negative rate (TNR).
Specificity equals the number of true negative voxels divided by the sum of true negative and false positive voxels. True negative and false positive values are computed on soft masks, see
"numeric_score".- Parameters:
prediction (ndarray) – First array.
groundtruth (ndarray) – Second array.
err_value (float) – Value returned in case of error.
- Returns:
Specificity score.
- Return type:
float
- intersection_over_union(prediction, groundtruth, err_value=0.0)[source]
Intersection of two (soft) arrays over their union (IoU).
- Parameters:
prediction (ndarray) – First array.
groundtruth (ndarray) – Second array.
err_value (float) – Value returned in case of error.
- Returns:
IoU.
- Return type:
float
- accuracy_score(prediction, groundtruth, err_value=0.0)[source]
Accuracy.
Accuracy equals the number of true positive and true negative voxels divided by the total number of voxels. True positive/negative and false positive/negative values are computed on soft masks, see
"numeric_score".- Parameters:
prediction (ndarray) – First array.
groundtruth (ndarray) – Second array.
- Returns:
Accuracy.
- Return type:
float
- multi_class_dice_score(im1, im2)[source]
Dice score for multi-label images.
Multi-class Dice score equals the average of the Dice score for each class. The first dimension of the input arrays is assumed to represent the classes.
- Parameters:
im1 (ndarray) – First array.
im2 (ndarray) – Second array.
- Returns:
Multi-class dice.
- Return type:
float
Postprocessing API
- nifti_capable(wrapped)[source]
Decorator to make a given function compatible with input being Nifti objects.
- Parameters:
wrapped – Given function.
- Returns:
Functions’ return.
- binarize_with_low_threshold(wrapped)[source]
Decorator to set low values (< 0.001) to 0.
- Parameters:
wrapped – Given function.
- Returns:
Functions’ return.
- multilabel_capable(wrapped)[source]
Decorator to make a given function compatible multilabel images.
- Parameters:
wrapped – Given function.
- Returns:
Functions’ return.
- threshold_predictions(predictions, thr=0.5)[source]
Threshold a soft (i.e. not binary) array of predictions given a threshold value, and returns a binary array.
- Parameters:
predictions (ndarray or nibabel object) – Image to binarize.
thr (float) – Threshold value: voxels with a value < to thr are assigned 0 as value, 1 otherwise.
- Returns:
ndarray or nibabel (same object as the input) containing only zeros or ones. Output type is int.
- Return type:
ndarray
- keep_largest_object(predictions)[source]
Keep the largest connected object from the input array (2D or 3D).
- Parameters:
predictions (ndarray or nibabel object) – Input segmentation. Image could be 2D or 3D.
- Returns:
ndarray or nibabel (same object as the input).
- keep_largest_object_per_slice(predictions, axis=2)[source]
Keep the largest connected object for each 2D slice, along a specified axis.
- Parameters:
predictions (ndarray or nibabel object) – Input segmentation. Image could be 2D or 3D.
axis (int) – 2D slices are extracted along this axis.
- Returns:
ndarray or nibabel (same object as the input).
- fill_holes(predictions, structure=(3, 3, 3))[source]
Fill holes in the predictions using a given structuring element. Note: This function only works for binary segmentation.
- Parameters:
predictions (ndarray or nibabel object) – Input binary segmentation. Image could be 2D or 3D.
structure (tuple of integers) – Structuring element, number of ints equals number of dimensions in the input array.
- Returns:
ndrray or nibabel (same object as the input). Output type is int.
- mask_predictions(predictions, mask_binary)[source]
Mask predictions using a binary mask: sets everything outside the mask to zero.
- Parameters:
predictions (ndarray or nibabel object) – Input binary segmentation. Image could be 2D or 3D.
mask_binary (ndarray) – Numpy array with the same shape as predictions, containing only zeros or ones.
- Returns:
ndarray or nibabel (same object as the input).
- coordinate_from_heatmap(nifti_image, thresh=0.3)[source]
Retrieve coordinates of local maxima in a soft segmentation. :param nifti_image: nifti image of the soft segmentation. :type nifti_image: nibabel object :param thresh: Relative threshold for local maxima, i.e., after normalizing :type thresh: float :param the min and max between 0 and 1: :param respectively.:
- Returns:
A list of computed coordinates found by local maximum. each element will be a list composed of [x, y, z]
- Return type:
list
- label_file_from_coordinates(nifti_image, coord_list)[source]
Creates a nifti object with single-voxel labels. Each label has a value of 1. The nifti object as the same orientation as the input. :param nifti_image: Path to the image which affine matrix will be used to generate a new image with :type nifti_image: nibabel object :param labels.: :param coord_list: list of coordinates. Each element is [x, y, z]. Orientation should be the same as the image :type coord_list: list
- Returns:
A nifti object containing the singe-voxel label of value 1. The matrix will be the same size as nifti_image.
- Return type:
nib_pred
- remove_small_objects(data, bin_structure, size_min)[source]
Removes all unconnected objects smaller than the minimum specified size.
- Parameters:
data (ndarray) – Input data.
bin_structure (ndarray) – Structuring element that defines feature connections.
size_min (int) – Minimal object size to keep in input data.
- Returns:
Array with small objects.
- Return type:
ndarray
- class Postprocessing(postprocessing_params, data_pred, dim_lst, filename_prefix)[source]
Bases:
objectPostprocessing steps manager
- Parameters:
postprocessing_params (dict) – Indicates postprocessing steps (in the right order)
data_pred (ndarray) – Prediction from the model.
dim_lst (list) – Dimensions of a voxel in mm.
filename_prefix (str) – Path to prediction file without suffix.
- Attributes:
postprocessing_params (dict) – Indicates postprocessing steps (in the right order)
data_pred (ndarray) – Prediction from the model.
px (float) – Resolution (mm) along the first axis.
py (float) – Resolution (mm) along the second axis.
pz (float) – Resolution (mm) along the third axis.
filename_prefix (str) – Path to prediction file without suffix.
n_classes (int) – Number of classes.
bin_struct (ndarray) – Binary structure.
- binarize_maxpooling()[source]
Binarize by setting to 1 the voxel having the max prediction across all classes.
- uncertainty(thr, suffix)[source]
Removes the most uncertain predictions.
- Parameters:
thr (float) – Uncertainty threshold.
suffix (str) – Suffix of uncertainty filename.
Testing API
- test(model_params, dataset_test, testing_params, path_output, device, cuda_available=True, metric_fns=None, postprocessing=None)[source]
Main command to test the network.
- Parameters:
model_params (dict) – Model’s parameters.
dataset_test (imed_loader) – Testing dataset.
testing_params (dict) – Testing parameters.
path_output (str) – Folder where predictions are saved.
device (torch.device) – Indicates the CPU or GPU ID.
cuda_available (bool) – If True, CUDA is available.
metric_fns (list) – List of metrics, see
ivadomed.metrics.postprocessing (dict) – Contains postprocessing steps.
- Returns:
result metrics.
- Return type:
dict
- run_inference(test_loader, model, model_params, testing_params, ofolder, cuda_available, i_monte_carlo=None, postprocessing=None)[source]
Run inference on the test data and save results as nibabel files.
- Parameters:
test_loader (torch DataLoader) –
model (nn.Module) –
model_params (dict) –
testing_params (dict) –
ofolder (str) – Folder where predictions are saved.
cuda_available (bool) – If True, CUDA is available.
i_monte_carlo (int) – i_th Monte Carlo iteration.
postprocessing (dict) – Indicates postprocessing steps.
- Returns:
Prediction, Ground-truth of shape n_sample, n_label, h, w, d.
- Return type:
ndarray, ndarray
- threshold_analysis(model_path, ds_lst, model_params, testing_params, metric='dice', increment=0.1, fname_out='thr.png', cuda_available=True)[source]
Run a threshold analysis to find the optimal threshold on a sub-dataset.
- Parameters:
model_path (str) – Model path.
ds_lst (list) – List of loaders.
model_params (dict) – Model’s parameters.
testing_params (dict) – Testing parameters
metric (str) – Choice between “dice” and “recall_specificity”. If “recall_specificity”, then a ROC analysis is performed.
increment (float) – Increment between tested thresholds.
fname_out (str) – Plot output filename.
cuda_available (bool) – If True, CUDA is available.
- Returns:
optimal threshold.
- Return type:
float
Training API
- train(model_params, dataset_train, dataset_val, training_params, path_output, device, wandb_params=None, cuda_available=True, metric_fns=None, n_gif=0, resume_training=False, debugging=False)[source]
Main command to train the network.
- Parameters:
model_params (dict) – Model’s parameters.
dataset_train (imed_loader) – Training dataset.
dataset_val (imed_loader) – Validation dataset.
training_params (dict) –
path_output (str) – Folder where log files, best and final models are saved.
device (str) – Indicates the CPU or GPU ID.
cuda_available (bool) – If True, CUDA is available.
metric_fns (list) – List of metrics, see
ivadomed.metrics.n_gif (int) – Generates a GIF during training if larger than zero, one frame per epoch for a given slice. The parameter indicates the number of 2D slices used to generate GIFs, one GIF per slice. A GIF shows predictions of a given slice from the validation sub-dataset. They are saved within the output path.
resume_training (bool) – Load a saved model (“checkpoint.pth.tar” in the path_output) for resume training. This training state is saved everytime a new best model is saved in the log directory.
debugging (bool) – If True, extended verbosity and intermediate outputs.
- Returns:
- best_training_dice, best_training_loss, best_validation_dice,
best_validation_loss.
- Return type:
float, float, float, float
- get_sampler(ds, balance_bool, metadata)[source]
Get sampler.
- Parameters:
ds (BidsDataset) – BidsDataset object.
balance_bool (bool) – If True, a sampler is generated that balance positive and negative samples.
- Returns:
Returns BalancedSampler, Bool: Sampler and boolean for shuffling (set to False). Otherwise: Returns None and True.
- Return type:
If balance_bool is True
- get_scheduler(params, optimizer, num_epochs=0)[source]
Get scheduler.
- Parameters:
params (dict) – scheduler parameters, see PyTorch documentation
optimizer (torch optim) –
num_epochs (int) – number of epochs.
- Returns:
torch.optim, bool, which indicates if the scheduler is updated for each batch (True), or for each epoch (False).
- get_loss_function(params)[source]
Get Loss function.
- Parameters:
params (dict) – See
ivadomed.losses.- Returns:
imed_losses object.
- get_metadata(metadata, model_params)[source]
Get metadata during batch loop.
- Parameters:
metadata (batch) –
model_params (dict) –
- Returns:
If FiLMedUnet, Returns a list of metadata, that have been transformed by the One Hot Encoder. If HeMISUnet, Returns a numpy array where each row represents a sample and each column represents a contrast.
- load_checkpoint(model, optimizer, gif_dict, scheduler, fname)[source]
Load checkpoint.
This function check if a checkpoint is available. If so, it updates the state of the input objects.
- Parameters:
model (nn.Module) – Init model.
optimizer (torch.optim) – Model’s optimizer.
gif_dict (dict) – Dictionary containing a GIF of the training.
scheduler (_LRScheduler) – Learning rate scheduler.
fname (str) – Checkpoint filename.
- Returns:
nn.Module, torch, dict, int, float, _LRScheduler, int
Transformations API
- multichannel_capable(wrapped)[source]
Decorator to make a given function compatible multichannel images.
- Parameters:
wrapped – Given function.
- Returns:
Functions’ return.
- two_dim_compatible(wrapped)[source]
Decorator to make a given function compatible 2D or 3D images.
- Parameters:
wrapped – Given function.
- Returns:
Functions’ return.
- class Compose(dict_transforms, requires_undo=False)[source]
Bases:
objectComposes transforms together.
Composes transforms together and split between images, GT and ROI.
- self.transform is a dict:
keys: “im”, “gt” and “roi”
values torchvision_transform.Compose objects.
- Attributes:
dict_transforms (dict) – Dictionary where the keys are the transform names and the value their parameters.
requires_undo (bool) – If True, does not include transforms which do not have an undo_transform implemented yet.
- Parameters:
transform (dict) – Keys are “im”, “gt”, “roi” and values are torchvision_transforms.Compose of the transformations of interest.
- class UndoCompose(compose)[source]
Bases:
objectUndo the Compose transformations.
Call the undo transformations in the inverse order than the “do transformations”.
- Attributes:
compose (torchvision_transforms.Compose)
- Parameters:
transforms (torchvision_transforms.Compose) –
- class UndoTransform(transform)[source]
Bases:
objectCall undo transformation.
- Attributes:
transform (ImedTransform)
- Parameters:
transform (ImedTransform) –
- class NumpyToTensor[source]
Bases:
ImedTransformConverts nd array to tensor object.
- class Resample(hspace, wspace, dspace=1.0)[source]
Bases:
ImedTransformResample image to a given resolution.
- Parameters:
hspace (float) – Resolution along the first axis, in mm.
wspace (float) – Resolution along the second axis, in mm.
dspace (float) – Resolution along the third axis, in mm.
interpolation_order (int) – Order of spline interpolation. Set to 0 for label data. Default=2.
- class NormalizeInstance[source]
Bases:
ImedTransformNormalize a tensor or an array image with mean and standard deviation estimated from the sample itself.
- class CroppableArray[source]
Bases:
ndarrayZero padding slice past end of array in numpy.
Adapted From: https://stackoverflow.com/a/41155020/13306686
- class Crop(size)[source]
Bases:
ImedTransformCrop data.
- Parameters:
size (tuple of int) – Size of the output sample. Tuple of size 2 if dealing with 2D samples, 3 with 3D samples.
- Attributes:
size (tuple of int) – Size of the output sample. Tuple of size 3.
- class ROICrop(size)[source]
Bases:
CropMake a crop of a specified size around a Region of Interest (ROI).
- class DilateGT(dilation_factor)[source]
Bases:
ImedTransformRandomly dilate a ground-truth tensor.
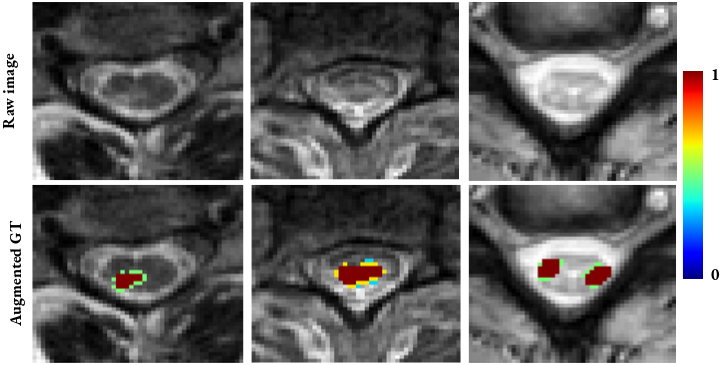
- Parameters:
dilation_factor (float) – Controls the number of dilation iterations. For each individual lesion, the number of
follows (dilation iterations is computed as) – nb_it = int(round(dilation_factor * sqrt(lesion_area)))
0 (If dilation_factor <=) –
performed. (then no dilation will be) –
- class RandomAffine(degrees=0, translate=None, scale=None)[source]
Bases:
ImedTransformApply Random Affine transformation.
- Parameters:
degrees (float) – Positive float or list (or tuple) of length two. Angles in degrees. If only a float is provided, then rotation angle is selected within the range [-degrees, degrees]. Otherwise, the list / tuple defines this range.
translate (list of float) – List of floats between 0 and 1, of length 2 or 3 depending on the sample shape (2D or 3D). These floats defines the maximum range of translation along each axis.
scale (list of float) – List of floats between 0 and 1, of length 2 or 3 depending on the sample shape (2D or 3D). These floats defines the maximum range of scaling along each axis.
- Attributes:
degrees (tuple of floats)
translate (list of float)
scale (list of float)
- class RandomReverse[source]
Bases:
ImedTransformMake a randomized symmetric inversion of the different values of each dimensions.
- class RandomShiftIntensity(shift_range, prob=0.1)[source]
Bases:
ImedTransformAdd a random intensity offset.
- Parameters:
shift_range (tuple of floats) – Tuple of length two. Specifies the range where the offset that is applied is randomly selected from.
prob (float) – Between 0 and 1. Probability of occurence of this transformation.
- class ElasticTransform(alpha_range, sigma_range, p=0.1)[source]
Bases:
ImedTransformApplies elastic transformation.
See also
Simard, Patrice Y., David Steinkraus, and John C. Platt. “Best practices for convolutional neural networks applied to visual document analysis.” Icdar. Vol. 3. No. 2003. 2003.
- Parameters:
alpha_range (tuple of floats) – Deformation coefficient. Length equals 2.
sigma_range (tuple of floats) – Standard deviation. Length equals 2.
- class AdditiveGaussianNoise(mean=0.0, std=0.01)[source]
Bases:
ImedTransformAdds Gaussian Noise to images.
- Parameters:
mean (float) – Gaussian noise mean.
std (float) – Gaussian noise standard deviation.
- class Clahe(clip_limit=3.0, kernel_size=(8, 8))[source]
Bases:
ImedTransformApplies Contrast Limited Adaptive Histogram Equalization for enhancing the local image contrast.
See also
Zuiderveld, Karel. “Contrast limited adaptive histogram equalization.” Graphics gems (1994): 474-485.
Default values are based on:
See also
Zheng, Qiao, et al. “3-D consistent and robust segmentation of cardiac images by deep learning with spatial propagation.” IEEE transactions on medical imaging 37.9 (2018): 2137-2148.
- Parameters:
clip_limit (float) – Clipping limit, normalized between 0 and 1.
kernel_size (tuple of int) – Defines the shape of contextual regions used in the algorithm. Length equals image
dimension (ie 2 or 3 for 2D or 3D, respectively) –
- class HistogramClipping(min_percentile=5.0, max_percentile=95.0)[source]
Bases:
ImedTransformPerforms intensity clipping based on percentiles.
- Parameters:
min_percentile (float) – Between 0 and 100. Lower clipping limit.
max_percentile (float) – Between 0 and 100. Higher clipping limit.
- class RandomGamma(log_gamma_range, p=0.5)[source]
Bases:
ImedTransformRandomly changes the contrast of an image by gamma exponential
- Parameters:
log_gamma_range (tuple of floats) – Log gamma range for changing contrast. Length equals 2.
p (float) – Probability of performing the gamma contrast
- class RandomBiasField(coefficients, order, p=0.5)[source]
Bases:
ImedTransformApplies a random MRI bias field artifact to the image via torchio.RandomBiasField()
- Parameters:
coefficients (float) – Maximum magnitude of polynomial coefficients
order – Order of the basis polynomial functions
p (float) – Probability of applying the bias field
- class RandomBlur(sigma_range, p=0.5)[source]
Bases:
ImedTransformApplies a random blur to the image
- Parameters:
sigma_range (tuple of floats) – Standard deviation range for the gaussian filter
p (float) – Probability of performing blur
- get_subdatasets_transforms(transform_params)[source]
Get transformation parameters for each subdataset: training, validation and testing.
- Parameters:
transform_params (dict) –
- Returns:
Training, Validation and Testing transformations.
- Return type:
dict, dict, dict
- get_preprocessing_transforms(transforms)[source]
Checks the transformations parameters and selects the transformations which are done during preprocessing only.
- Parameters:
transforms (dict) – Transformation dictionary.
- Returns:
Preprocessing transforms.
- Return type:
dict
- apply_preprocessing_transforms(transforms, seg_pair, roi_pair=None) Tuple[dict, dict][source]
Applies preprocessing transforms to segmentation pair (input, gt and metadata).
- Parameters:
transforms (Compose) – Preprocessing transforms.
seg_pair (dict) – Segmentation pair containing input and gt.
roi_pair (dict) – Segementation pair containing input and roi.
- Returns:
Segmentation pair and roi pair.
- Return type:
tuple
- prepare_transforms(transform_dict, requires_undo=True)[source]
This function separates the preprocessing transforms from the others and generates the undo transforms related.
- Parameters:
transform_dict (dict) – Dictionary containing the transforms and there parameters.
requires_undo (bool) – Boolean indicating if transforms can be undone.
- Returns:
- transform lst containing the preprocessing transforms and regular transforms, UndoCompose
object containing the transform to undo.
- Return type:
list, UndoCompose
- tio_transform(x, transform)[source]
Applies TorchIO transformations to a given image and returns the transformed image and history.
- Parameters:
x (np.ndarray) – input image
transform (tio.transforms.Transform) – TorchIO transform
- Returns:
transformed image, history of parameters used for the applied transformation
- Return type:
np.ndarray, list
Utils API
- class Metavar(value)[source]
Bases:
EnumThis class is used to display intuitive input types via the metavar field of argparse.
- initialize_wandb(wandb_params)[source]
Initializes WandB and based upon the parameters sets it up or disables it for experimental tracking
- Parameters:
wandb_params (dict) – wandb parameters
- Returns:
True if wandb tracking is enabled
- Return type:
bool, wandb_tracking
- cuda(input_var, cuda_available=True, non_blocking=False)[source]
Passes input_var to GPU.
- Parameters:
input_var (Tensor) – either a tensor or a list of tensors.
cuda_available (bool) – If False, then return identity
non_blocking (bool) –
- Returns:
Tensor
- unstack_tensors(sample)[source]
Unstack tensors.
- Parameters:
sample (Tensor) –
- Returns:
list of Tensors.
- Return type:
list
- generate_sha_256(context: dict, df, file_lst: List[str]) None[source]
generate sha256 for a training file
- Parameters:
context (dict) – configuration context.
df (pd.DataFrame) – Dataframe containing all BIDS image files indexed and their metadata.
file_lst (List[str]) – list of strings containing training files
- save_onnx_model(model, inputs, model_path)[source]
Convert PyTorch model to ONNX model and save it as model_path.
- Parameters:
model (nn.Module) – PyTorch model.
inputs (Tensor) – Tensor, used to inform shape and axes.
model_path (str) – Output filename for the ONNX model.
- define_device(gpu_id)[source]
Define the device used for the process of interest.
- Parameters:
gpu_id (int) – GPU ID.
- Returns:
True if cuda is available.
- Return type:
Bool, device
- display_selected_model_spec(params)[source]
Display in terminal the selected model and its parameters.
- Parameters:
params (dict) – Keys are param names and values are param values.
- display_selected_transfoms(params, dataset_type)[source]
Display in terminal the selected transforms for a given dataset.
- Parameters:
params (dict) –
dataset_type (list) – e.g. [‘testing’] or [‘training’, ‘validation’]
- plot_transformed_sample(before, after, list_title=None, fname_out='', cmap='jet')[source]
Utils tool to plot sample before and after transform, for debugging.
- Parameters:
before (ndarray) – Sample before transform.
after (ndarray) – Sample after transform.
list_title (list of str) – Sub titles of before and after, resp.
fname_out (str) – Output filename where the plot is saved if provided.
cmap (str) – Matplotlib colour map.
- check_exe(name)[source]
Ensure that a program exists.
- Parameters:
name (str) – Name or path to program.
- Returns:
path of the program or None
- Return type:
str or None
- get_arguments(parser, args)[source]
Get arguments from function input or command line.
- Parameters:
parser (argparse.ArgumentParser) – ArgumentParser object
args (list) – either a list of arguments or None. The list should be formatted like this: [“-d”, “SOME_ARG”, “–model”, “SOME_ARG”]
- format_path_data(path_data)[source]
- Parameters:
path_data (list or str) – Either a list of paths, or just one path.
- Returns:
A list of paths
- Return type:
list
- similarity_score(a: str, b: str) float[source]
use DiffLIb SequenceMatcher to resolve the similarity between text. Help make better choice in terms of derivatives. :param a: a string :param b: another string
Returns: a score indicative of the similarity between the sequence.
- get_timestamp() str[source]
Return a datetime string in the format YYYY-MM-DDTHHMMSS.(sub-precision) Returns:
- get_win_system_memory() float[source]
Obtain the amount of memory available on Windows system. Returns: memory in GB Source: https://stackoverflow.com/a/21589439
- get_linux_system_memory() float[source]
Obtain the amount of memory available on Linux system. Returns: memory in GB Source: https://stackoverflow.com/a/28161352
- get_mac_system_memory() float[source]
Obtain the amount of memory available on MacOS system. Returns: memory in GB Source: https://apple.stackexchange.com/a/4296
Visualize API
- overlap_im_seg(img, seg)[source]
Overlap image (background, greyscale) and segmentation (foreground, jet).
- class LoopingPillowWriter(fps=5, metadata=None, codec=None, bitrate=None)[source]
Bases:
PillowWriter
- class AnimatedGif(size)[source]
Bases:
objectGenerates GIF.
- Parameters:
size (tuple) – Size of frames.
- Attributes:
fig (plt)
size_x (int)
size_y (int)
images (list) – List of frames.
- save_color_labels(gt_data, binarize, gt_filename, output_filename, slice_axis)[source]
Saves labels encoded in RGB in specified output file.
- Parameters:
gt_data (ndarray) – Input image with dimensions (Number of classes, height, width, depth).
binarize (bool) – If True binarizes gt_data to 0 and 1 values, else soft values are kept.
gt_filename (str) – GT path and filename.
output_filename (str) – Name of the output file where the colored labels are saved.
slice_axis (int) – Indicates the axis used to extract slices: “axial”: 2, “sagittal”: 0, “coronal”: 1.
- Returns:
RGB labels.
- Return type:
ndarray
- convert_labels_to_RGB(grid_img)[source]
Converts 2D images to RGB encoded images for display in tensorboard.
- Parameters:
grid_img (Tensor) – GT or prediction tensor with dimensions (batch size, number of classes, height, width).
- Returns:
RGB image with shape (height, width, 3).
- Return type:
tensor
- save_img(writer, epoch, dataset_type, input_samples, gt_samples, preds, wandb_tracking=False, is_three_dim=False)[source]
Saves input images, gt and predictions in tensorboard (and wandb depending upon the inputs in the config file).
- Parameters:
writer (SummaryWriter) – Tensorboard’s summary writer.
epoch (int) – Epoch number.
dataset_type (str) – Choice between Training or Validation.
input_samples (Tensor) – Input images with shape (batch size, number of channel, height, width, depth) if 3D else (batch size, number of channel, height, width)
gt_samples (Tensor) – GT images with shape (batch size, number of channel, height, width, depth) if 3D else (batch size, number of channel, height, width)
preds (Tensor) – Model’s prediction with shape (batch size, number of channel, height, width, depth) if 3D else (batch size, number of channel, height, width)
is_three_dim (bool) – True if 3D input, else False.
- save_feature_map(batch, layer_name, path_output, model, test_input, slice_axis)[source]
Save model feature maps.
- Parameters:
batch (dict) –
layer_name (str) –
path_output (str) – Output folder.
model (nn.Module) – Network.
test_input (Tensor) –
slice_axis (int) – Indicates the axis used for the 2D slice extraction: Sagittal: 0, Coronal: 1, Axial: 2.
- class HookBasedFeatureExtractor(submodule, layername, upscale=False)[source]
Bases:
ModuleThis function extracts feature maps from given layer. Helpful to observe where the attention of the network is focused.
https://github.com/ozan-oktay/Attention-Gated-Networks/tree/a96edb72622274f6705097d70cfaa7f2bf818a5a
- Parameters:
submodule (nn.Module) – Trained model.
layername (str) – Name of the layer where features need to be extracted (layer of interest).
upscale (bool) – If True output is rescaled to initial size.
- Attributes:
submodule (nn.Module) – Trained model.
layername (str) – Name of the layer where features need to be extracted (layer of interest).
outputs_size (list) – List of output sizes.
outputs (list) – List of outputs containing the features of the given layer.
inputs (list) – List of inputs.
inputs_size (list) – List of input sizes.
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- __init__(submodule, layername, upscale=False)[source]
Initialize internal Module state, shared by both nn.Module and ScriptModule.
- forward(x)[source]
Define the computation performed at every call.
Should be overridden by all subclasses.
Note
Although the recipe for forward pass needs to be defined within this function, one should call the
Moduleinstance afterwards instead of this since the former takes care of running the registered hooks while the latter silently ignores them.
Inference API
- onnx_inference(model_path: str, inputs: tensor) tensor[source]
Run ONNX inference
- Parameters:
model_path (str) – Path to the ONNX model.
inputs (Tensor) – Batch of input image.
- Returns:
Network output.
- Return type:
Tensor
- get_preds(context: dict, fname_model: str, model_params: dict, cuda_available: bool, device: device, batch: dict) tensor[source]
Returns the predictions from the given model.
- Parameters:
context (dict) – configuration dict.
fname_model (str) – name of file containing model.
model_params (dict) – dictionary containing model parameters.
cuda_available (bool) – True if cuda is available.
device (torch.device) – Device used for prediction.
batch (dict) – dictionary containing input, gt and metadata
- Returns:
predictions from the model.
- Return type:
tensor
- get_onehotencoder(context: dict, folder_model: str, options: dict, ds: Dataset) dict[source]
Returns one hot encoder which is needed to update the model parameters when FiLMedUnet is applied.
- Parameters:
context (dict) – Configuration dict.
folder_model (str) – Foldername which contains trained model and its configuration file.
options (dict) – Contains film metadata information.
ds (Dataset) – Dataset used for the segmentation.
- Returns:
onehotencoder used in the model params.
- Return type:
dict
- pred_to_nib(data_lst: List[ndarray], z_lst: List[int], fname_ref: str, fname_out: str, slice_axis: int, debug: bool = False, kernel_dim: str = '2d', bin_thr: float = 0.5, discard_noise: bool = True, postprocessing: dict | None = None) Nifti1Image[source]
Save the network predictions as nibabel object.
Based on the header of fname_ref image, it creates a nibabel object from the Network predictions (data_lst).
- Parameters:
data_lst (list of np arrays) – Predictions, either 2D slices either 3D patches.
z_lst (list of ints) – Slice indexes to reconstruct a 3D volume for 2D slices.
fname_ref (str) – Filename of the input image: its header is copied to the output nibabel object.
fname_out (str) – If not None, then the generated nibabel object is saved with this filename.
slice_axis (int) – Indicates the axis used for the 2D slice extraction: Sagittal: 0, Coronal: 1, Axial: 2.
debug (bool) – If True, extended verbosity and intermediate outputs.
kernel_dim (str) – Indicates whether the predictions were done on 2D or 3D patches. Choices: ‘2d’, ‘3d’.
bin_thr (float) – If positive, then the segmentation is binarized with this given threshold. Otherwise, a soft segmentation is output.
discard_noise (bool) – If True, predictions that are lower than 0.01 are set to zero.
postprocessing (dict) – Contains postprocessing steps to be applied.
- Returns:
NiBabel object containing the Network prediction.
- Return type:
nibabel.Nifti1Image
- pred_to_png(pred_list: list, target_list: list, subj_path: str, suffix: str = '', max_value: int = 1)[source]
Save the network predictions as PNG files with suffix “_target_pred”.
- Parameters:
pred_list (list of np arrays) – list of 2D predictions.
target_list (list of str) – list of target suffixes.
subj_path (str) – Path of the subject filename in output folder without extension (e.g. “path_output/pred_masks/sub-01_sample-01_SEM”).
suffix (str) – additional suffix to append to the filename (e.g. “_pred.png”)
max_value (int) – Maximum mask value of the float mask to use during the conversion to uint8.
- process_transformations(context: dict, fname_roi: str, fname_prior: str, metadata: dict, slice_axis: int, fname_images: list) dict[source]
Sets the transformation based on context parameters. When ROI is not provided center-cropping is applied.
If there is an object_detection_path, then we modify the metadata to store transformation data.
- Parameters:
context (dict) – configuration dictionary.
fname_roi (str) – filename containing region for cropping image prior to segmentation.
fname_prior (str) – prior image filename.
metadata (dict) – metadata used in setting bounding box when we have object_detection_params.
slice_axis (int) – Indicates the axis used for the 2D slice extraction: Sagittal: 0, Coronal: 1, Axial: 2.
fname_images (list) – list of image filenames (e.g. .nii.gz) to segment.
- Returns:
metadata.
- Return type:
dict
- set_option(options: dict, postpro: dict, context: dict, key: str)[source]
Generalized function that sets postprocessing option based on given list of options.
When given key already exists in options, we initialize the key value for the postprocessing dictionary Otherwise, when the key is already found in the postprocessing attritute of the context, we remove it
- Parameters:
options (dict) – Contains postprocessing steps information.
postpro (dict) – Postprocessing settings.
context (dict) – Configuration dict.
key (str) – The key of the postprocessing option we wish to set.
- Returns:
postprocessing settings.
- Return type:
dict
- set_postprocessing_options(options: dict, context: dict)[source]
Updates the postprocessing options based on existing settings found in options.
- Parameters:
options (dict) – Contains postprocessing steps information.
context (dict) – Configuration dict.
- segment_volume(folder_model: str, fname_images: list, gpu_id: int = 0, options: dict | None = None)[source]
Segment an image.
Segment an image (fname_image) using a pre-trained model (folder_model). If provided, a region of interest (fname_roi) is used to crop the image prior to segment it.
- Parameters:
folder_model (str) – foldername which contains (1) the model (‘folder_model/folder_model.pt’) to use (2) its configuration file (‘folder_model/folder_model.json’) used for the training, see https://github.com/neuropoly/ivadomed/wiki/configuration-file
fname_images (list) – list of image filenames (e.g. .nii.gz) to segment. Multichannel models require multiple images to segment, e.i., len(fname_images) > 1.
gpu_id (int) – Number representing gpu number if available. Currently does NOT support multiple GPU segmentation.
options (dict) –
This can optionally contain any of the following key-value pairs:
’binarize_prediction’: (float) Binarize segmentation with specified threshold. Predictions below the threshold become 0, and predictions above or equal to threshold become 1. Set to -1 for no thresholding (i.e., soft segmentation).
’binarize_maxpooling’: (bool) Binarize by setting to 1 the voxel having the maximum prediction across all classes. Useful for multiclass models.
’fill_holes’: (bool) Fill small holes in the segmentation.
’keep_largest’: (bool) Keep the largest connected-object for each class from the output segmentation.
’remove_small’: (list of str) Minimal object size to keep with unit (mm3 or vox). A single value can be provided or one value per prediction class. Single value example: [“1mm3”], [“5vox”]. Multiple values example: [“10”, “20”, “10vox”] (remove objects smaller than 10 voxels for class 1 and 3, and smaller than 20 voxels for class 2).
’pixel_size’: (list of float) List of microscopy pixel size in micrometers. Length equals 2 [PixelSizeX, PixelSizeY] for 2D or 3 [PixelSizeX, PixelSizeY, PixelSizeZ] for 3D, where X is the width, Y the height and Z the depth of the image.
’pixel_size_units’: (str) Units of pixel size (Must be either “mm”, “um” or “nm”)
’no_patch’: (bool) 2D patches are not used while segmenting with models trained with patches. The “no_patch” option supersedes the “overlap_2D” option. This option may not be suitable with large images depending on computer RAM capacity.
’overlap_2D’: (list of int) List of overlaps in pixels for 2D patching. Length equals 2 [OverlapX, OverlapY], where X is the width and Y the height of the image.
’metadata’: (str) Film metadata.
’fname_prior’: (str) An image filename (e.g., .nii.gz) containing processing information (e.g., spinal cord segmentation, spinal location or MS lesion classification, spinal cord centerline), used to crop the image prior to segment it if provided. The segmentation is not performed on the slices that are empty in this image.
- Returns:
List of nibabel objects containing the soft segmentation(s), one per prediction class, List of target suffix associated with each prediction in pred_list
- Return type:
list, list
- split_classes(nib_prediction)[source]
Split a 4D nibabel multi-class segmentation file in multiple 3D nibabel binary segmentation files.
- Parameters:
nib_prediction (nibabelObject) – 4D nibabel object.
- Returns:
list of nibabelObject.
- reconstruct_3d_object(context: dict, batch: dict, undo_transforms: UndoCompose, preds: tensor, preds_list: list, kernel_3D: bool, is_2d_patch: bool, slice_axis: int, slice_idx_list: list, data_loader: DataLoader, fname_images: list, i_batch: int, last_sample_bool: bool, weight_matrix: tensor, volume: tensor, image: tensor)[source]
Reconstructs the 3D object from the current batch, and returns the list of predictions and targets.
- Parameters:
context (dict) – configuration dict.
batch (dict) – Dictionary containing input, gt and metadata
undo_transforms (UndoCompose) – Undo transforms so prediction match original image resolution and shape
preds (tensor) – Subvolume predictions
preds_list (list of tensor) – list of subvolume predictions.
kernel_3D (bool) – true when using 3D kernel.
is_2d_patch (bool) – Indicates if 2d patching is used.
slice_axis (int) – Indicates the axis used for the 2D slice extraction: Sagittal: 0, Coronal: 1, Axial: 2.
slice_idx_list (list of int) – list of indices for the axis slices.
data_loader (DataLoader) – DataLoader object containing batches using in object construction.
fname_images (list) – list of image filenames (e.g. .nii.gz) to segment.
i_batch (int) – index of current batch.
last_sample_bool (bool) – flag to indicate whether this is the last sample in the 3D volume
weight_matrix (tensor) – the weight matrix
volume (tensor) – the volume tensor that is being partially reconstructed through the loop
image (tensor) – the image tensor that is being partially reconstructed through the loop
- Returns:
list of predictions target_list (list): list of targets last_sample_bool (bool): flag to indicate whether this is the last sample in the 3D volume weight_matrix (tensor): the weight matrix. Must be returned as passing tensor by reference is NOT reliable. volume (tensor): the volume tensor that is being partially reconstructed through the loop. Must be returned as passing tensor by reference is NOT reliable. image (tensor): the vimage tensor that is being partially reconstructed through the loop. Must be returned as passing tensor by reference is NOT reliable.
- Return type:
pred_list (list)
- volume_reconstruction(batch: dict, pred: tensor, undo_transforms: UndoCompose, smp_idx: int, volume: tensor | None = None, weight_matrix: tensor | None = None)[source]
Reconstructs volume prediction from subvolumes used during training :param batch: Dictionary containing input, gt and metadata :type batch: dict :param pred: Subvolume prediction :type pred: tensor :param undo_transforms: Undo transforms so prediction match original image resolution and shap :type undo_transforms: UndoCompose :param smp_idx: Batch index :type smp_idx: int :param volume: Reconstructed volume :type volume: tensor :param weight_matrix: Weights containing the number of predictions for each voxel :type weight_matrix: tensor
- Returns:
undone subvolume, metadata (dict): metadata, last_sample_bool (bool): boolean representing if its the last sample of the volume volume (tensor): representing the volume reconstructed weight_matrix (tensor): weight matrix
- Return type:
pred_undo (tensor)
- image_reconstruction(batch: dict, pred: tensor, undo_transforms: UndoCompose, smp_idx: int, image: tensor | None = None, weight_matrix: tensor | None = None)[source]
Reconstructs image prediction from patches used during training :param batch: Dictionary containing input, gt and metadata :type batch: dict :param pred: Patch prediction :type pred: tensor :param undo_transforms: Undo transforms so prediction match original image resolution and shape :type undo_transforms: UndoCompose :param smp_idx: Batch index :type smp_idx: int :param image: Reconstructed image :type image: tensor :param weight_matrix: Weights containing the number of predictions for each pixel :type weight_matrix: tensor
- Returns:
undone image metadata (dict): metadata last_patch_bool (bool): boolean representing if its the last patch of the image image (tensor): representing the image reconstructed weight_matrix (tensor): weight matrix
- Return type:
pred_undo (tensor)
Mixup API
- mixup(data, targets, alpha, debugging=False, ofolder=None)[source]
Compute the mixup data.
See also
Zhang, Hongyi, et al. “mixup: Beyond empirical risk minimization.” arXiv preprint arXiv:1710.09412 (2017).
- Parameters:
data (Tensor) – Input images.
targets (Tensor) – Input masks.
alpha (float) – MixUp parameter.
debugging (Bool) – If True, then samples of mixup are saved as png files.
ofolder (str) – If debugging, output folder where “mixup” folder is created and samples are saved.
- Returns:
Mixed image, Mixed mask.
- Return type:
Tensor, Tensor
- save_mixup_sample(ofolder, input_data, labeled_data, lambda_tensor)[source]
Save mixup samples as png files in a “mixup” folder.
- Parameters:
ofolder (str) – Output folder where “mixup” folder is created and samples are saved.
input_data (Tensor) – Input image.
labeled_data (Tensor) – Input masks.
lambda_tensor (Tensor) –
Uncertainty API
- run_uncertainty(image_folder)[source]
Compute uncertainty from model prediction.
This function loops across the model predictions (nifti masks) and estimates the uncertainty from the Monte Carlo samples. Both voxel-wise and structure-wise uncertainty are estimates.
- Parameters:
image_folder (str) – Folder containing the Monte Carlo samples.
- combine_predictions(fname_lst, fname_hard, fname_prob, thr=0.5)[source]
Combine predictions from Monte Carlo simulations.
- Combine predictions from Monte Carlo simulations and save the resulting as:
fname_prob, a soft segmentation obtained by averaging the Monte Carlo samples.
fname_hard, a hard segmentation obtained thresholding with thr.
- Parameters:
fname_lst (list of str) – List of the Monte Carlo samples.
fname_hard (str) – Filename for the output hard segmentation.
fname_prob (str) – Filename for the output soft segmentation.
thr (float) – Between 0 and 1. Used to threshold the soft segmentation and generate the hard segmentation.
- voxelwise_uncertainty(fname_lst, fname_out, eps=1e-05)[source]
Estimate voxel wise uncertainty.
Voxel-wise uncertainty is estimated as entropy over all N MC probability maps, and saved in fname_out.
- Parameters:
fname_lst (list of str) – List of the Monte Carlo samples.
fname_out (str) – Output filename.
eps (float) – Epsilon value to deal with np.log(0).
- structurewise_uncertainty(fname_lst, fname_hard, fname_unc_vox, fname_out)[source]
Estimate structure wise uncertainty.
Structure-wise uncertainty from N MC probability maps (fname_lst) and saved in fname_out with the following suffixes:
‘-cv.nii.gz’: coefficient of variation
‘-iou.nii.gz’: intersection over union
‘-avgUnc.nii.gz’: average voxel-wise uncertainty within the structure.
- Parameters:
fname_lst (list of str) – List of the Monte Carlo samples.
fname_hard (str) – Filename of the hard segmentation, which is used to compute the avgUnc by providing a mask of the structures.
fname_unc_vox (str) – Filename of the voxel-wise uncertainty, which is used to compute the avgUnc.
fname_out (str) – Output filename.
Maths API
- rescale_values_array(arr, minv=0.0, maxv=1.0, dtype=<class 'numpy.float32'>)[source]
Rescale the values of numpy array arr to be from minv to maxv.
- Parameters:
arr (ndarry) – Array whose values will be rescaled.
minv (float) – Minimum value of the output array.
maxv (float) – Maximum value of the output array.
dtype (type) – Cast array to this type before performing the rescaling.
- gaussian_kernel(kernlen=10)[source]
Create a 2D gaussian kernel with user-defined size.
- Parameters:
kernlen (int) – size of kernel
- Returns:
a 2D array of size (kernlen,kernlen)
- Return type:
ndarray
- heatmap_generation(image, kernel_size)[source]
Generate heatmap from image containing sing voxel label using convolution with gaussian kernel :param image: 2D array containing single voxel label :type image: ndarray :param kernel_size: size of gaussian kernel :type kernel_size: int
- Returns:
2D array heatmap matching the label.
- Return type:
ndarray